Data Quality Control
High-throughput sequencing techniques have become the leading method to study, decode and discover the genomic origins of biological phenomenons. EGA provides a secure archival of such identifiable genomics data with the purpose of data-upcycling, i.e. to re-use these data for research. High-quality data standards are essential to ensure the quality and credibility of the research. Moreover, a quality check report can assure a researcher beforehand about the data that they will request access, therefore saving time and effort.
The EGA has developed a File Quality Control Report (QC Report) to provide generic quality control reports for Fastq, SAM/BAM/CRAM, and VCF files deposited at EGA. This QC Report will allow users to get information regarding the files submitted within a specific dataset. The data requesters will obtain information such as the quality of reads, mapped reads, number of variants, and other features before starting the requesting process, which will save the efforts and time.
Accessing file quality control reports
In each dataset page, the user can explore the files that it contains by clicking the "files" tab.
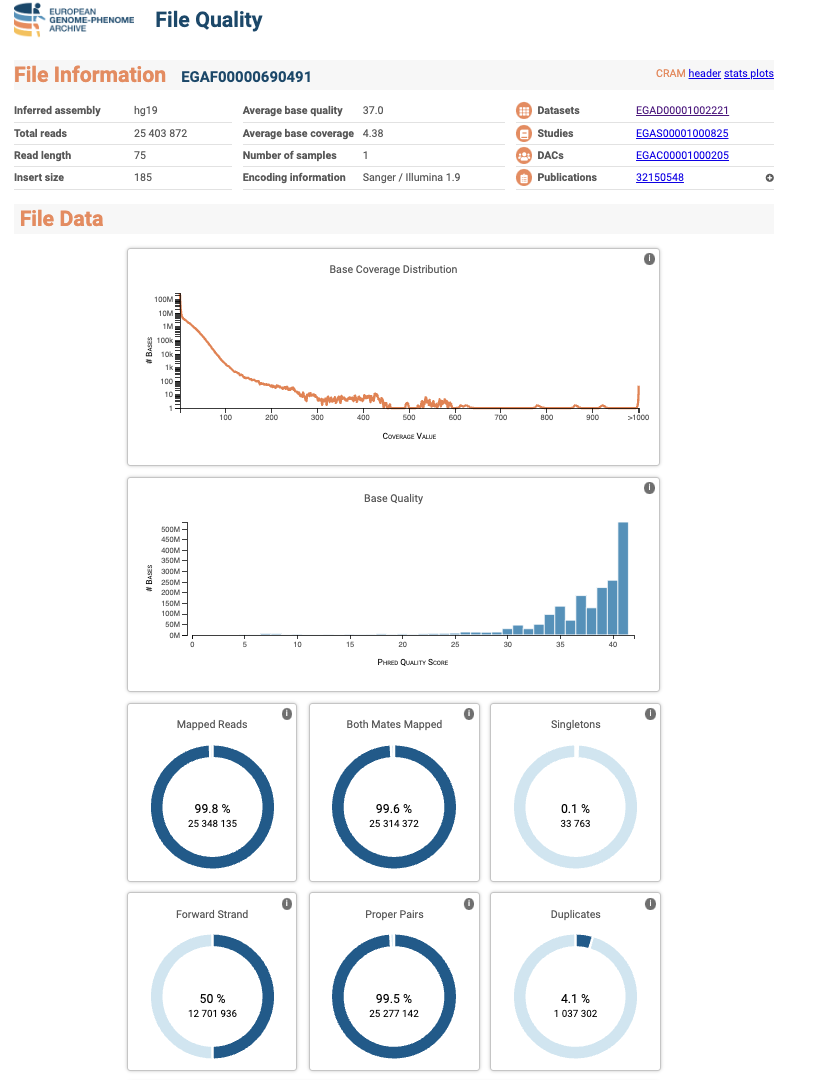
The Quality Control report of a file has two sections. The first one, contains general information about the file, such as the inferred assembly, total reads, the dataset or study where it comes from, etc. The second section contains plots that summarise interesting information about the file, for example, the base coverage distribution, base quality or mapped reads.
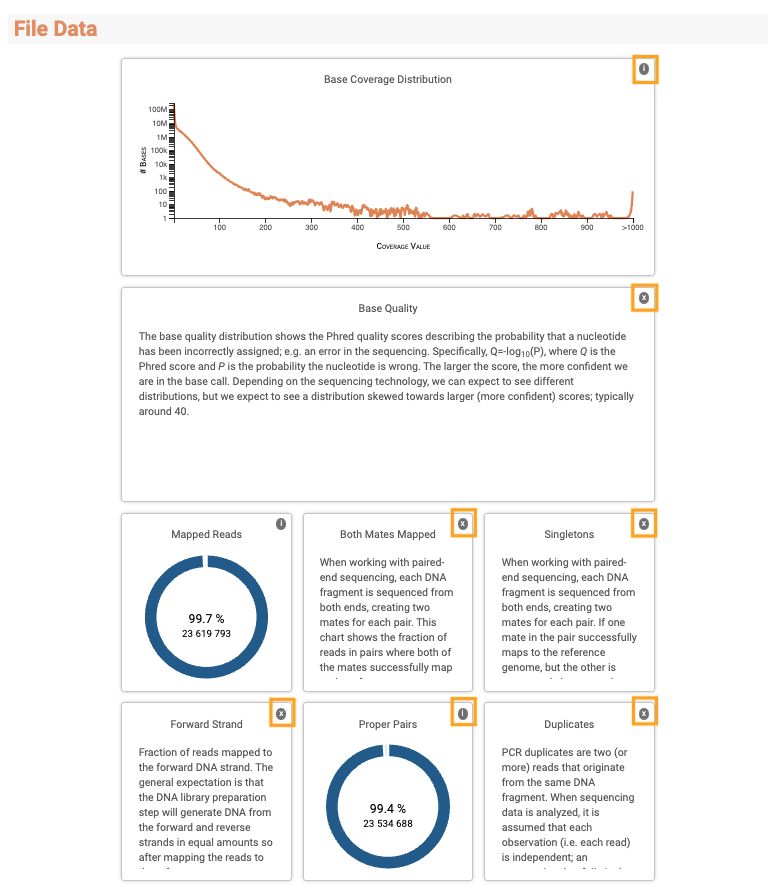
The description of each plot is accessible by clicking the "i" button at the top-right corner of each plot box.
Technical Description
For analysing the fastq, SAM/BAM/CRAM and VCF files, the EGA applies a set of tools widely used in the bioinformatics community.
- FASTQ: FastQC, recognised as the gold standard tool by the community.
- Per base sequence quality, per sequence quality scores, per base sequence content, per sequence GC content, sequence duplication levels, etc.
- SAM/BAM/CRAM: samtools, also the gold standard, generates results plots useful to get an overall idea of the quality of the file.
- base coverage distribution, base quality, % of mapped reads, % of both mates mapped, singletons, duplicates, etc.
- VCF: vcftools and bcftools, combined with a custom script to infer the genome assembly.
- site frequency distribution, Ts/Tv, base changes, indel distribution, etc.
